Objective
This study demonstrates the use of Nanoxis LPI™ FlowCell in conjunction with LC-MS/MS to profile the protein content of a plasma membrane preparation derived from Arabidopsis thaliana, with respect to identity, membrane association types and subcellular locations.
Method
Arabidopsis plasma membranes were isolated using an aqueous two-phase system1. LPI™ Maxi FlowCells were used to immobilize plasma membrane vesicles which were then digested with trypsin to produce peptide samples suitable for LC-MS/MS analysis. Plasma membranes were isolated and donated by the laboratory of Plant Physiology of the University of Groningen, the Netherlands.
Results
Peptide matching using Mascot searching against the Swiss-Prot Arabidopsis thaliana subset resulted in identification of 249 proteins based on 843 unique peptides.
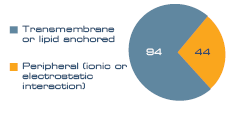
To assess the membrane purity of the sample a membrane association analysis was performed. 138 proteins (55%) were classed as membrane associated. A further 22% were noted as ribosomal, which can be classified as both soluble and membrane bound, and the remaining 23% were classed as soluble. Within the membrane protein group, proteins anchored to the membrane by a transmembrane domain or lipid anchor dominated (94), compared to peripheral (44), as seen in Figure 1.
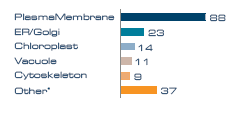
Figure 2. Proteins per subcellular annotation.
* Nucleus, cytosol, mitochondria, peroxisome and ribosome.
Subcellular localization analysis of the membrane proteins revealed that the dominant membrane compartment represented was the plasma membrane, indicating that the aqueous two-phase enrichment process used for the sample preparation was efficient. 88 membrane proteins originated from the plasma membrane, with smaller contributions from ER/Golgi, vacuole, chloroplast and cytoskeleton (Figure 2). Note that some proteins have dual location and are therefore counted twice in this graph.
Visit Download Center for detailed report >>
References
1. Larsson, C., Widell, S., Kjellbom, P. (1987) Methods Enzymol. 148, 558-568
Download Center
Download applications data, brochures, publications and more
Visit Download Center >>
Contact
Ordering
order@nanoxis.com
Technical service
techservice@nanoxis.com Nanoxis AB
MC2 Building A
Kemivägen 9
SE - 412 96 Gothenburg
Sweden
info@nanoxis.com
Distributors >>